6 Lesser-Known Scikit-Learn Features That Will Save You Time
For many people studying data science, Scikit-Learn is often the first machine learning library they encounter. It’s because Scikit-Learn offers various APIs that are useful for model development while still being easy for beginners to use.
As helpful as they may be, many features from Scikit-Learn are rarely explored and have untapped potential. This article will explore six lesser-known features that will save you time.
1. Validation Curve
The first function we will explore is the validation curve function from Scikit-Learn. From the name, you can guess that it performs some kind of validation, but it’s not just simple validation that is performs. The function explores machine learning model performance over various values of specific hyperparameters.
Using the cross-validation method, the validation curve function evaluates training and test performance over the range of hyperparameter values. The process results in two sets of scores that we can compare visually.
Let’s try out the function with sample data and visualize the results. First, let’s load sample data and set up the hyperparameter range we want to explore. In this case, we will explore how the SVC model’s accuracy performance over various gamma hyperparameters.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 |
import numpy as np import matplotlib.pyplot as plt from sklearn.model_selection import validation_curve from sklearn.svm import SVC from sklearn.datasets import load_iris
data = load_iris() X, y = data.data, data.target
param_range = np.logspace(–3, 3, 5)
train_scores, test_scores = validation_curve( SVC(), X, y, param_name=“gamma”, param_range=param_range, cv=5, scoring=“accuracy” ) |
Once you execute the code above, you will get two scores: train_scores & test_scores.
These are both assigned arrays of scores like below.
|
(array([[0.925 , 0.925 , 0.93333333, 0.925 , 0.9 ], [0.975 , 0.94166667, 0.975 , 0.96666667, 0.95833333], [0.975 , 0.98333333, 0.99166667, 0.99166667, 0.99166667], [1. , 1. , 1. , 1. , 1. ], [1. , 1. , 1. , 1. , 1. ]]), array([[0.86666667, 0.96666667, 0.83333333, 0.96666667, 0.93333333], [0.93333333, 0.96666667, 0.93333333, 0.93333333, 1. ], [0.96666667, 1. , 0.9 , 0.96666667, 1. ], [0.86666667, 0.73333333, 0.7 , 0.8 , 0.9 ], [0.46666667, 0.4 , 0.33333333, 0.4 , 0.4 ]])) |
We can visualize the validation curve using code similar to the following.
|
plt.figure(figsize=(8, 6)) plt.plot(param_range, train_mean, label=“Training score”, color=“blue”, marker=“o”) plt.plot(param_range, test_mean, label=“Cross-validation score”, color=“green”, marker=“s”) plt.xscale(“log”) plt.xlabel(“Gamma”) plt.ylabel(“Accuracy”) plt.title(“Validation Curve for SVC (gamma parameter)”) plt.legend(loc=“best”) plt.show() |
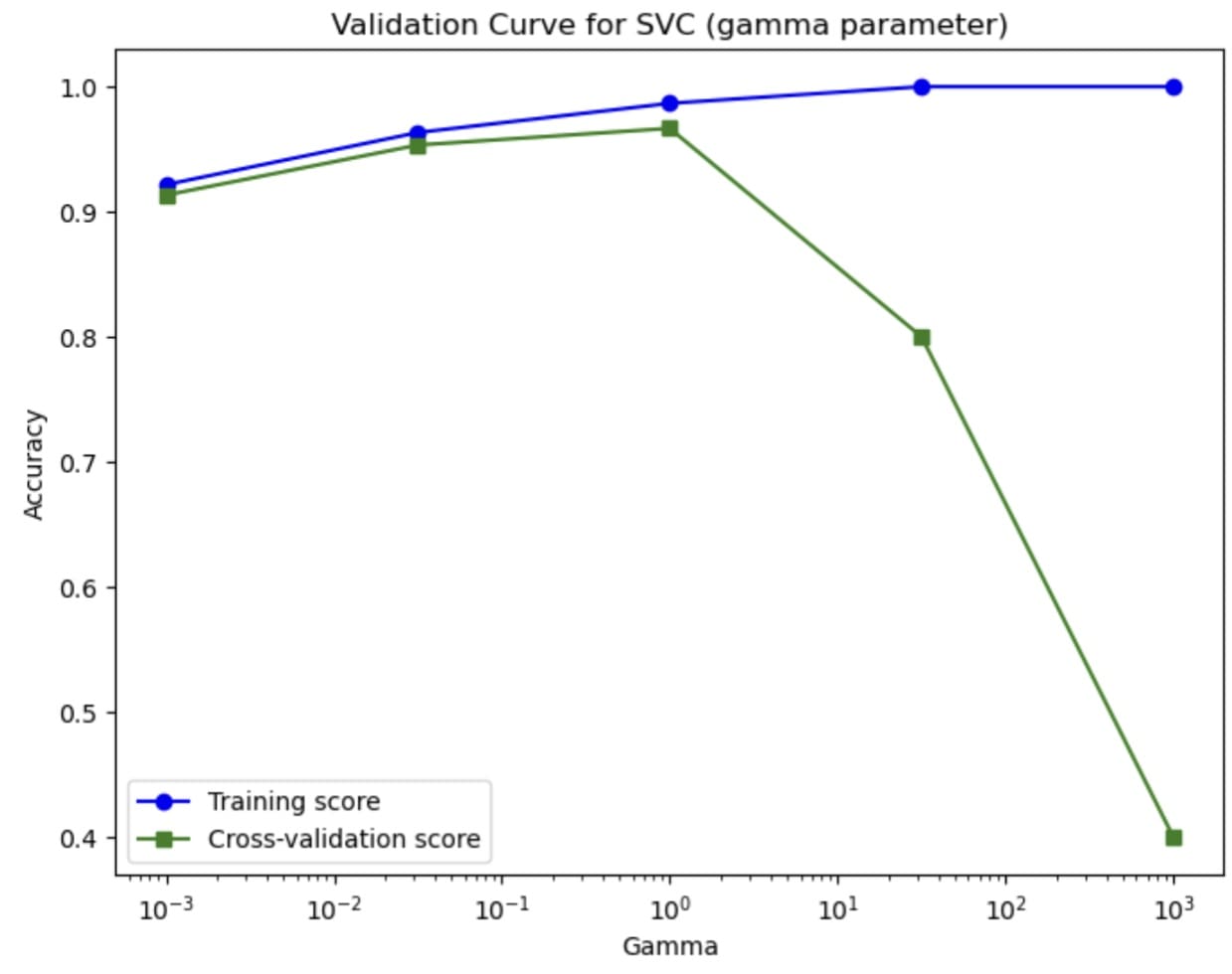
The curve teaches us how the hyperparameters affect the model’s performance. Using the validation curve, we can find the optimal value for the hyperparameter and estimate it better than relying on the simple train-test split.
Try using a validation curve in your model development process to guide you in developing the best model possible and avoid issues such as overfitting.
2. Model Calibration
When we develop a machine learning classifier model, we need to remember that it’s not enough simply to provide correct classification prediction; the probabilities associated with the prediction must also be reliable. The process to ensure that the probabilities are reliable is called calibration.
The calibration process adjusts the model’s probability estimation. The technique pushes the probability to reflect the true likelihood of the prediction so it is not overconfident or underconfident. The uncalibrated model might predict an event with a 90% probability chance, while the actual success rate is much lower, which means the model was overconfident. That’s why we need to calibrate the model.
By calibrating the model, we could improve the trust in model prediction and inform the user of the actual estimation of the actual risk from the model.
Let’s try the calibration process with Scikit-Learn. The library offers a diagnostic function (calibration_curve) and a model calibration class (CalibratedClassifierCV).
We will use breast cancer data and the logistic regression model as the basis. Then, we will compare the original model with the calibrated model for the probability.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 |
import numpy as np import matplotlib.pyplot as plt from sklearn.datasets import load_breast_cancer from sklearn.linear_model import LogisticRegression from sklearn.calibration import calibration_curve, CalibratedClassifierCV from sklearn.model_selection import train_test_split
data = load_breast_cancer() X, y = data.data, data.target
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=42) lr = LogisticRegression().fit(X_train, y_train)
prob_pos_lr = lr.predict_proba(X_test)[:, 1] fraction_lr, mean_pred_lr = calibration_curve(y_test, prob_pos_lr, n_bins=10)
calibrated_clf = CalibratedClassifierCV(lr, cv=‘prefit’, method=‘isotonic’) calibrated_clf.fit(X_train, y_train) prob_pos_calibrated = calibrated_clf.predict_proba(X_test)[:, 1] fraction_cal, mean_pred_cal = calibration_curve(y_test, prob_pos_calibrated, n_bins=10)
plt.figure(figsize=(8, 6)) plt.plot(mean_pred_lr, fraction_lr, marker=‘o’, label=‘Original LR’) plt.plot(mean_pred_cal, fraction_cal, marker=‘s’, label=‘Calibrated LR (Isotonic)’) plt.plot([0, 1], [0, 1], linestyle=‘–‘, label=‘Perfect Calibration’) plt.xlabel(“Mean predicted probability”) plt.ylabel(“Fraction of positives”) plt.title(“Calibration Curve Comparison”) plt.legend(loc=“upper left”) plt.show() |

We can see that the calibrated logistic regression is closer to the model with perfect calibration than the original. This means that the calibrated model can better estimate the actual risk, although it is still not ideal.
Try using the calibration method to improve the model prediction capability.
3. Permutation Importance
Whenever we work with a machine learning model, we use the data features to provide the prediction result. However, not every feature contributes to the prediction in the same manner.
The permutation_importance() method is for measuring the feature contribution to model performances by randomly permuting (changing) feature values and evaluating the model performance after the permutation. If the model performance degrades, the feature impacts the model; conversely, if the model performance is unchanged, it suggests that the feature might not be that useful for the specific model performance.
The technique is straightforward and intuitive, making it helpful in interpreting any model’s internal decision-making. It’s beneficial for models with no inherent feature importance method embedded inside.
Let’s try out the model with a Python code example. We will use sample data and models similar to our previous example.
|
import numpy as np import matplotlib.pyplot as plt from sklearn.datasets import load_iris from sklearn.linear_model import LogisticRegression from sklearn.inspection import permutation_importance from sklearn.model_selection import train_test_split
data = load_iris() X, y = data.data, data.target
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=42)
model = LogisticRegression() model.fit(X_train, y_train)
result = permutation_importance(model, X_test, y_test, n_repeats=10, random_state=42, scoring=‘accuracy’) |
With the code above, we have the model and permutation importance result, where we will analyze the feature’s impact on the model. Let’s look at the average and standard deviation results for permutation importance.
|
feature_names = data.feature_names importances = result.importances_mean std = result.importances_std
for i, name in enumerate(feature_names): print(f“{name}: Mean importance = {importances[i]:.4f} (+/- {std[i]:.4f})”) |
The result is as follows.
|
sepal length (cm): Mean importance = 0.0132 (+/– 0.0177) sepal width (cm): Mean importance = 0.0000 (+/– 0.0000) petal length (cm): Mean importance = 0.6000 (+/– 0.0805) petal width (cm): Mean importance = 0.1553 (+/– 0.0362) |
We can also visualize the result above to understand the model performance better.
|
plt.barh(feature_names, importances, xerr=std) plt.xlabel(“Decrease in accuracy”) plt.title(“Permutation Importance”) plt.show() |
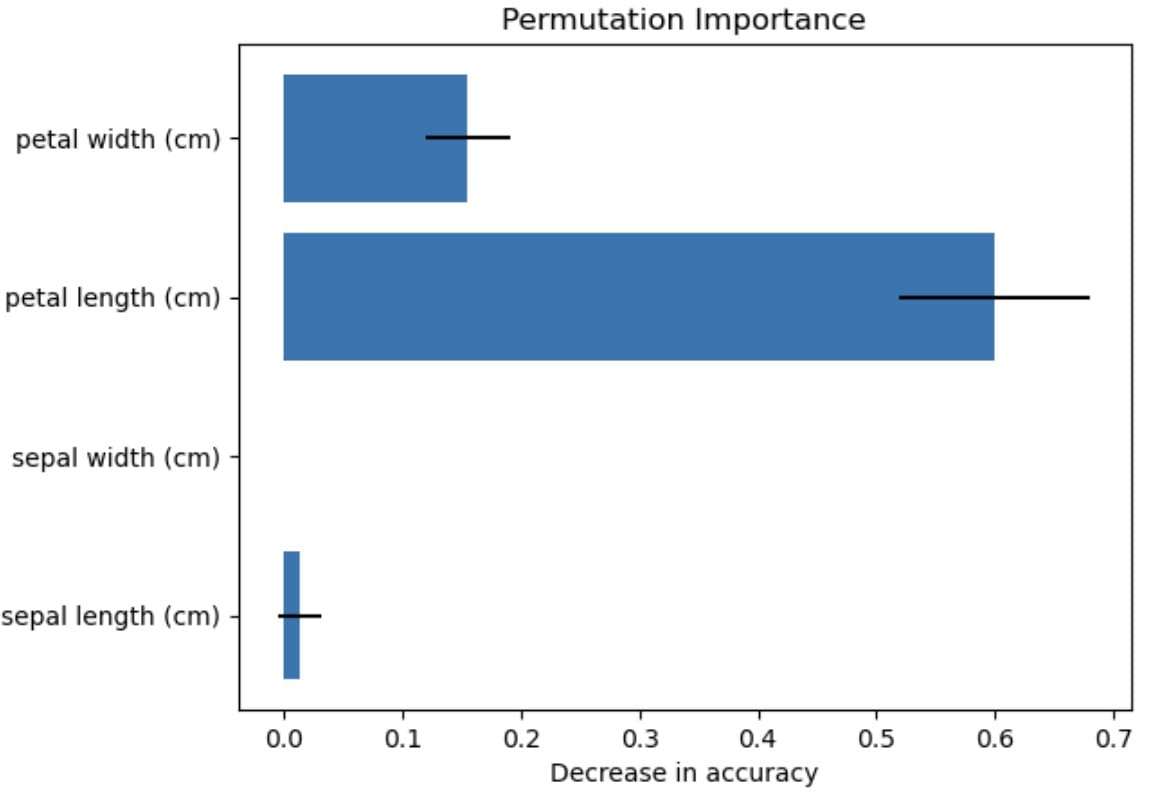
The visualization shows that the petal length most impacts the feature performance, while the sepal width has no effect. There is always uncertainty, which is represented by the standard deviation, but we can conclude with the permutation importance technique that petal length has the most impact.
That’s a simple feature importance technique that you can use for your next project.
4. Feature Hasher
Working on features for data science modeling, I often found that high-dimensional features were too memory intensive, which impacted the application’s overall performance. There are many ways to improve performance, such as dimensionality reduction or feature selection. The hashing method is another method that might be rarely used but could be helpful.
Hashing is converting data into a sparse numeric matrix with a fixed size. Applying a hash function to each feature can map the represented feature into a sparse matrix. We will use a hash function via FeatureHasher to compute the matrix column corresponding to a name.
Let’s try it out with the Python code.
|
import pandas as pd import seaborn as sns from sklearn.feature_extraction import FeatureHasher
titanic = sns.load_dataset(“titanic”)
titanic_sample = titanic[[‘sex’, ’embarked’, ‘class’]].dropna()
data_dicts = titanic_sample.to_dict(orient=‘records’) hasher = FeatureHasher(n_features=10, input_type=‘dict’)
hashed_features = hasher.transform(data_dicts)
hashed_array = hashed_features.toarray() print(“\nHashed feature matrix (dense format):\n”, hashed_array) |
You will see the output looks like the one below.
|
Hashed feature matrix (dense format): [[ 1. 0. 0. ... 0. –1. 0.] [–1. 0. 0. ... 0. 0. 0.] [ 1. 0. 0. ... 0. 0. 0.] ... [ 1. 0. 0. ... 0. 0. 0.] [–1. 0. 0. ... 0. –1. 0.] [ 1. 0. 0. ... 0. –1. 0.]] |
The dataset has been represented into a sparse matrix with 10 different features. You can specify the number of hash features you need to balance the memory usage and information loss.
5. Robust Scaler
Real-world data is rarely clean, and more often than not riddled with outliers. While an outlier is not intrinsically bad and might give information that contributes to the actual insight, there are times when it will skew our model results.
There are many techniques for scaling our outliers, but sometimes they can introduce bias. That’s why robust scaling is important to help preprocess our data. Robust scaling transforms the data by removing the median and scaling them according to the IQR instead of using mean and standard deviation.
The robust scaler is proper, with only a few outliers at extreme positions. By applying it, the dataset is stable and not influenced much by the outliers, which makes it useful for any machine learning model development.
Here is an example of using the robust scaler. Let’s use the Iris data example and introduce an outlier in the dataset.
|
import numpy as np import pandas as pd from sklearn.datasets import load_iris from sklearn.preprocessing import robust_scale import matplotlib.pyplot as plt
iris = load_iris() X = iris.data
outlier = np.array([[10, 10, 10, 10]]) X_out = np.vstack([X, outlier])
X_scaled = robust_scale(X_out) |
The above code easily scales the data with our introduced outlier. Try using yourself if you are having a hard time with outliers.
6. Feature Union
Feature union is a Scikit-Learn feature that combines multiple feature transformations within the pipeline. Instead of transforming the same features sequentially, feature union inputs the data into several transformers simultaneously to provide all the transformed features.
It’s a helpful feature where different transformers are required to capture various aspects of data and need to be present in the dataset. One transformer might used for the PCA technique, while the others use robust scaling.
Let’s try it out with the following code below. For example, we can create transformers for both PCA and polynomial features transformers.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 |
import numpy as np from sklearn.datasets import load_iris from sklearn.decomposition import PCA from sklearn.preprocessing import PolynomialFeatures from sklearn.pipeline import FeatureUnion
data = load_iris() X, y = data.data, data.target
pca = PCA(n_components=2)
poly = PolynomialFeatures(degree=2, include_bias=False)
union = FeatureUnion(transformer_list=[ (‘pca’, pca), (‘poly’, poly) ])
X_transformed = union.fit_transform(X) |
The example result of the transformed features is shown in the output below.
|
[–2.68412563 0.31939725 5.1 3.5 1.4 0.2 26.01 17.85 7.14 1.02 12.25 4.9 0.7 1.96 0.28 0.04 ] |
The result contains the PCA and polynomial features we have previously transformed.
Try experimenting with several transformations to see if they suit your analysis.
Conclusion
Scikit-Learn is a library that many data scientists use to develop models easily from their data. It’s easy to use and provides many features that are useful for our model work, yet many of those features seem underutilized though they could save you time.
In this article, we have explored six of these underutilized features, from validation curves to feature hashing to a robust scaler that isn’t highly influenced by outliers. Hopefully you found something new and of value herein, and I hope this has helped!

